Studies on the pathogenicity factors and development of simple methods for identification of plant pathogenic bacteria
A. Determination of pathogenicity factors of plant pathogenic Xanthomonas campestris.
We first reported that some cruciferous seed lots were infested with Xanthomonas campestris pv. campestris (XCC), some with X. campestris pv. raphani (XCR), and some with non-pathogenic xanthomonads. Based on the analyses of whole genome sequence, it was found that non-pathogenic X. campestris E1 (Xan-E1) isolate contains no entire pathogenicity island (PI; hrp gene cluster) and all type III effector protein genes existing in the pathogenic XCC and XCR. The Xan-E1 could acquire the entire pathogenicity island from the endemic XCC and XCR strains through the conjugation, but type III effector genes were not co-transferred. The studies first showed that the non-pathogenic X. campestris indeed exist on the cruciferous seeds, but it could be differentiated by the PCR assays using the genes of PI and type III effector as makers. Nevertheless, the non-pathogenic X. campestris might be a potential gene resource to cause the genetic diversity in the endemic pathogenic XCC and XCR through the gene transfer.
B. Development of the simple methods for identification of plant pathogenic bacteria
B-1. A Xan-D differential medium was developed for the isolation of Xanthomonas campestris pv. campestris, a bacterial black rot pathogen of crucifers
A xanthomonad differential medium (designated Xan-D medium) was developed, on which streaks and colonies of xanthomonads turned wet-shining yellow-green, and were surrounded with a smaller milky and a bigger clear zones in 3-4 days. The characteristics could easily be differentiated from yellow non-xanthomonads and other bacteria. The Xan-D medium was used to isolate and differentiate Xanthomonas campestris pv. campestris from naturally infected cabbages with black rot symptoms for a rapid diagnosis. The mechanism of color change and formation of a milky zone on the medium is mainly due to the Tween 80 hydrolytic capacity of xanthomonads. The gene, estA, responsible for Tween 80 hydrolysis was cloned and expressed in E. coli, which acquired a capacity to hydrolyze Tween 80, and could turn green and form a milky zone on the Xan-D medium.
B-2. The Xan-immunostrip was developed to rapid identification of X. campestris pv. campestris (XCC).
Detection of XCC on crucifer seeds is one of important quarantine measures in many countries. XCC differential and selective medium (Xan-D medium) was developed. In addition, polyclonal antibodies of XCC were also prepared amd used to develop the immunostrip for identification of XCC. The standard operational protocol of seed detection is to combine the Xan-D medium with the developed immunostrip to increase the sensitivity and specificity.
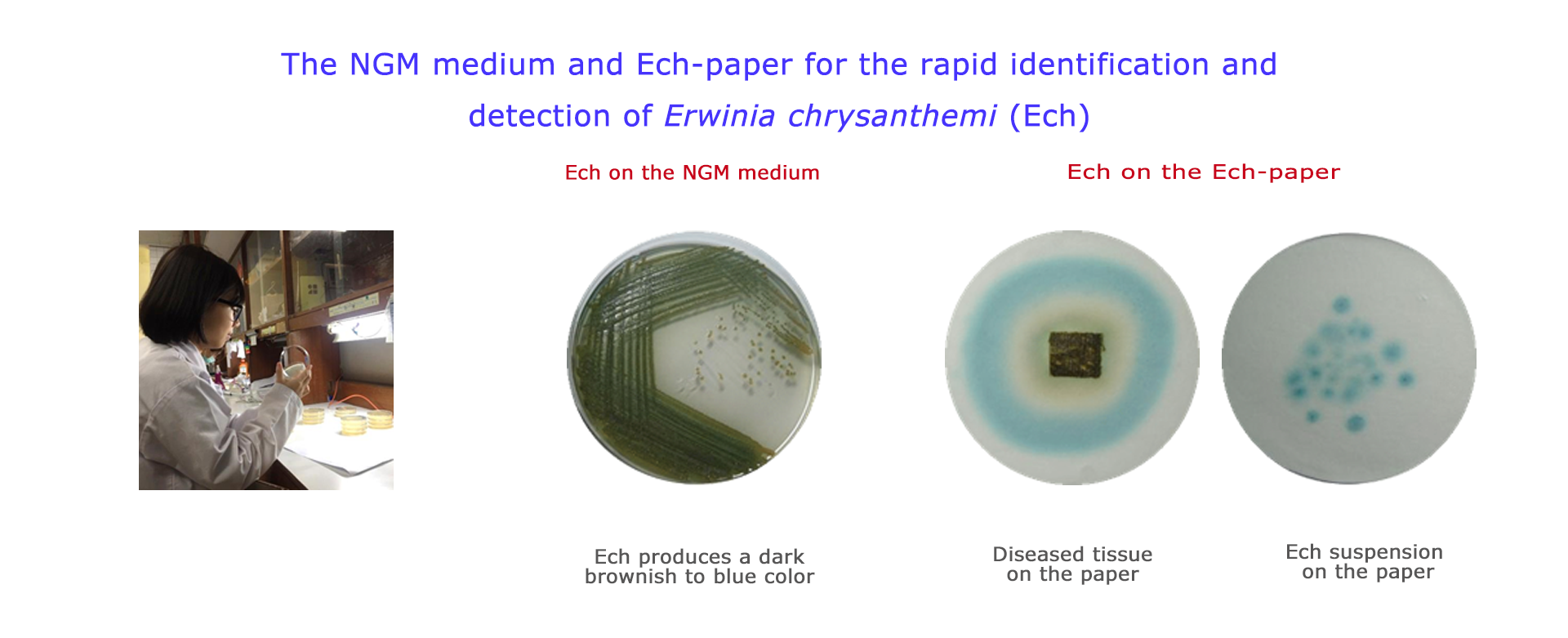
B-3. The NGM medium was developed for rapid identification of Dickeya chrysanthemi (syn. Erwinia chrysanthemi)
The NGM medium was developed for the isolation and differentiation of E. chrysanthemi from other Erwinia spp. based on the production of blue-pigmented indigoidine. The medium, named NGM, consists of nutrient agar supplemented with 1% glycerol, that induces pigment production, and 2mM MnCl2.4H2O, that further enhances color development. All tested strains of E. chrysanthemi grew well on the NGM medium, developing dark brownish to blue colonies easily distinguishable from other Erwinia spp. The results indicate that pigment production on the NGM medium is a very stable property and can be used as a phenotypic property to differentiate E. chrysanthemi from other Erwinia spp.
In addition, the NGM medium can also be used for differential isolation of E. chrysanthemi, Burkholderia gladioli, and B. glumae, which are the causal agents of leaf rot of Phalaenopsis and Oncidium orchids at three cultivation areas in Taiwan. E. chrysanthemi developed blue colonies, and all B. gladioli and B. glumae strains tested produced diffusible yellow pigments on the NGM medium, easily distinguishable from other Erwinia spp. and Burkholderia spp.
B-4. The Ech-paper was developed for the rapid diagnosis and identification of Dickeya chrysanthemi (syn. Erwinia chrysanthemi)
On the Ech-paper, the bacterial suspension of E. chrysanthemi and the plant tissue infected with E. chrysanthemi will turn blue in 1-2 days. The Ech-paper is an efficient method to identify E. chrysanthemi, and also a diagnosis method to determine whether the plant tissues are infected with E. chrysanthemi.
C. Identification and molecular typing of plant bacterial pathogens in Taiwan
C-1. Identification and molecular typing of Ralstonia solanacearum race 1, a bacterial wilt pathogen of solanaceous crops
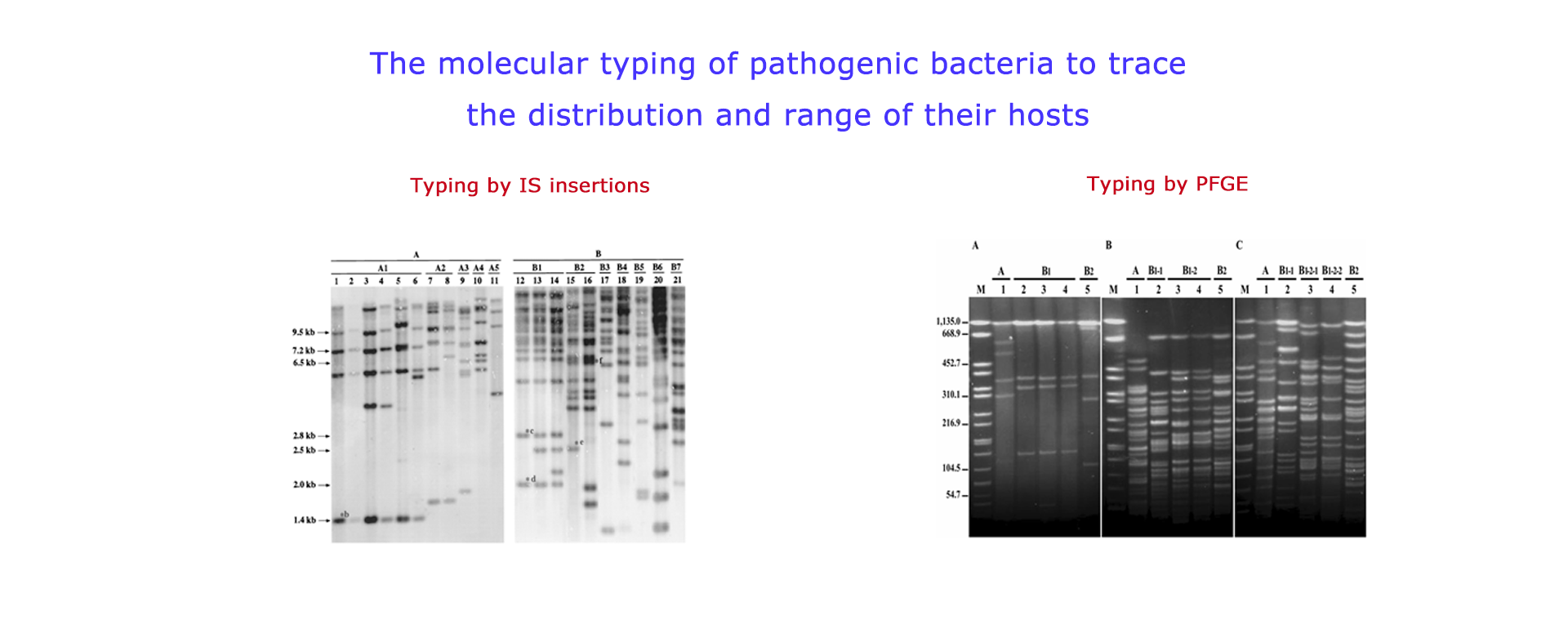
A new insertion sequence (IS), IS1405, was isolated and characterized from a Ralstonia solanacearum race 1 strain. Nucleotide sequence of IS1405 was used to design specific oligonucleotide primers for detection of race 1 strains by PCR. The PCR amplified a specific DNA fragment for all R. solanacearum race 1 strains tested, and no amplification was observed with some other plant pathogenic bacteria. RFLP pattern analysis using IS1405 as a probe revealed extensive genetic variation among strains of R. solanacearum race 1 isolated from 8 different host plants in Taiwan, which allows for tracking different subgroup strains of R. solanacearum through a host plant community. Furthermore, specific insertion sites of IS1405 in certain subgroups were used as a genetic marker to develop subgroup-specific primers for detection of R. solanacearum.
C-2. Identification and molecular typing of banana finger-tip rot pathogen
The pathogen of banana finger-tip rot was identified as Burkholderia cenocepacia (genomovar III of B. cepacia complex), which is a common plant-associated bacterium but also an important opportunistic pathogen of humans. Genetic diversity was assessed using recA PCR-restriction fragment length polymorphism (RFLP), recA nucleotide sequence analysis, and pulsed-field gel electrophoresis assays. PCR assays were further used to determine whether B. cenocepacia from bananas contain the cable pilus subunit gene (cblA), IS1356, and B. cepacia epidemic strain marker (BCESM), which are DNA markers associated with epidemic B. cepacia clinic strains. The results indicated that cblA and IS1356 were absent but the BCESM was found in all isolates. The present study revealed that banana is a natural reservoir of genetically diversified B. cenocepacia strains.
C-3. Identification of new bacterial diseases in Taiwan
The causal pathogens of following diseases are first identified in the Lab:
(1) The bacterial leaf blight of syngonium caused by Xanthomonas campestris pv. syngonii in Taiwan
(2) The bacterial leaf spot of poinsettia caused by Xanthomonas axonopodis pv. poinsettiicola in Taiwan
(3) The bacterial leaf blight of coriander caused by Xanthomonas campestris pv. coriandri in Taiwan.
(4) The bacterial leaf blight of white flowered calla lily caused by Xanthomonas campestris pv. zantedeschiae in Taiwan
(5) The bacterial soft rot of white flowered calla lily caused by Erwinia chrysanthemi in Taiwan
115 views